Getting started with R
Installation
First of all:
- Install R from the web site Rproject. R is a open-source (i.e. free) software.
- Install Rstudio from the web site RStudio. RStudio allows the user to run R in a more user-friendly environment. It is a open-source (i.e. free) software.
RStudio screen is divided in several windows.
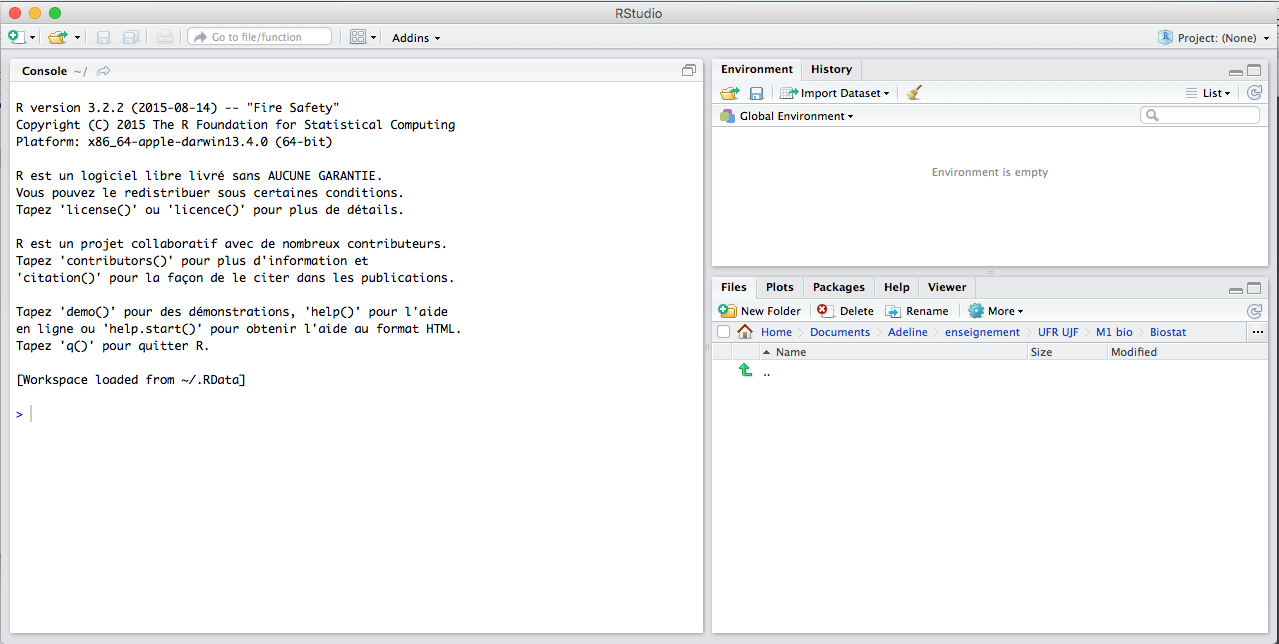
Alt text
- The console (lower left corner) is where you can type the commands and see output. Run the commands with Enter.
- The script (upper left corner) is where you can write the 'script' to save all your commands. Run them down to the lower left corner by Ctrl+R (Windows) or Cmd+Enter (Mac).
- The workspace (upper right corner) tab shows all the active objects. The workspace tab stores any object, value, function or anything you create during your R session. The {} tab shows a list of commands used so far. The history tab keeps a record of all previous commands.
- The files (lower right corner) tab show all the files and folders in your default workspace as if you were on a PC/Mac window. The plots tab will list a series of packages or add-ons needed to run certain processes. For additional info, see the help tab.
If you have different projects, we recommend to create a folder for each project. Then you can change the R working directory using the function setwd:
R> getwd() # Shows the working directory (wd) R> setwd("/M1 bio/Biostat") # Changes the wd
A few commands
An object can be created with the "assign" operator which is written as an arrow with a bracket and a minus sign.
R> n <- 10
One of the simplest commands is to type the name of an object to display its content.
R> n [1] 10
The digit 1 within brackets indicates that the display starts at the first element of n.
R is case sensitive (lower case different from capital letters).
R> x <-1 R> X <-10 R> x [1] 1 R> X [1] 10
Note that you can type an expression without assigning its value to an object, the result is displayed on the console but not stored in memory:
R> (10 + 2) * 5 [1] 60
The function ls lists the objects in memory.
R> ls() [1] "A" "D" "D1" "Dyndoc.Vec" [5] "dynRbVar" "dynVar" "dynVarWithArg" "E" [9] "Fo" "freq" "G1" "G2" [13] "Hy" "HY" "LenzI" "LenzT" [17] "M" "ms" "mu" "muC" [21] "muR" "n" "N" "p" [25] "pstatus" "pstatusadB" "pstatusadBH" "pstatusadBY" [29] "q" "R" "R2" "S" [33] "sample" "sdC" "sdf" "sdR" [37] "sds" "sig" "SS" "Tmu1" [41] "TRS" "x" "X" "Y"
The on-line help of R gives useful information on how to use the function. Help is available directly for a given function. For example:
R> ? hist
displays the help page for the function hist() (histogram).
Data and objects
Objects
Objects in R are characterized by two attributes which specify the kind of data represented by an object. In order to understand the usefulness of these attributes, consider a variable that takes the value 1, 2, or 3: such a variable could be an integer variable (for instance, the number of eggs in a nest), or the coding of a categorical variable (for instance, grade of cancer).
It is clear that the statistical analysis of this variable will not be the same in both cases: with R, the attributes of the object give the necessary information.
All objects have two attributes: mode and length. The mode is the basic type of the elements of the object; there are four main modes: numeric, character, complex, and logical (FALSE or TRUE). The length is the number of elements of the object. To display the mode and the length of an object, one can use the functions mode and length, respectively:
R> x<-1 R> mode(x) [1] "numeric" R> length(x) [1] 1
Whatever the mode, missing data are represented by NA (not available).
Reading data in a file
For reading and writing in files, R uses the working directory. To find this directory, the command getwd() (get working directory) can be used, and the working directory can be changed with setwd("C:/data"). It is necessary to give the path to a file if it is not in the working directory.
Data can be read with the function read.table or scan. The function read.table creates a data frame. For instance, a file named datafile.csv can be read:
R> mydata <- read.table("data/datafile.csv")
In that command, mydata is the name you choose for the data frame. By defaults, each variable of the data frame is named V1, V2, .... They can be accessed individually by mydata$V1, mydata$V2, ... or by mydata["V1"], mydata["V2"], ... or by mydata[, 1], mydata[, 2], ....
All the options of the function read.table are described in the help file. For example, if the file contains the names of the variables on its first line, we can use the option header, if the cells are separated by ; , we can use the option sep=";":
R> XY <- read.table("data/datafile.csv",header=TRUE,sep=";")
Example To upload the data set called hypoxy.csv, run the following instruction:
R> HY <- read.table("data/hypoxy.csv", header=TRUE, dec=",")
To check if the data were correctly loaded, use the function head that displays the first 6 rows of the dataset:
R> head(HY) Level Name_Prot Hypoxy Training N_Rat Location 1 0.9843 RyR2 No No N1 TA 2 0.9419 RyR2 No No N2 TA 3 0.7761 RyR2 No No N5 TA 4 0.8668 RyR2 No No N7 TA 5 1.2249 RyR2 No No N9 TA 6 1.2061 RyR2 No No N10 TA
All the values of the first six rows are displayed, the first is numeric, the five others are not.
Generating data
Example: to create the vector (1,2,3,4,5), use the command
R> 1:5 [1] 1 2 3 4 5
The resulting vector has 5 elements. Arithmetic operators can be used:
R> 1:5-1 [1] 0 1 2 3 4 R> 1:(5-1) [1] 1 2 3 4
Example: create a vector with 9 equispaced numbers between 1 and 5:
R> seq(1, 5, 0.5) [1] 1.0 1.5 2.0 2.5 3.0 3.5 4.0 4.5 5.0
Example: add a value 6 to the vector X equal to (1,2,3,4,5):
R> X <- 1:5 R> c(X,6) [1] 1 2 3 4 5 6
Example: create a vector with 10 elements equal to 1:
R> rep(1, 10) [1] 1 1 1 1 1 1 1 1 1 1
Example: create a vector with 1 repeated 5 times, 2 repeated 5 times and 3 repeated 5 times:
R> gl(3, 5) [1] 1 1 1 1 1 2 2 2 2 2 3 3 3 3 3 Levels: 1 2 3
Example: Create first the vector Y <- c(1,4,9, 16, 25). Check the length of Y and bind the two vectors X and Y:
R> Y <- c(1,4,9, 16, 25) R> cbind(X,Y) X Y [1,] 1 1 [2,] 2 4 [3,] 3 9 [4,] 4 16 [5,] 5 25
Example: Bind the two vectors X and Y in row:
R> rbind(X,Y) [,1] [,2] [,3] [,4] [,5] X 1 2 3 4 5 Y 1 4 9 16 25
Example: Create a matrix with elements from 1 to 6, with 2 rows and 3 columns:
R> A <- matrix(1:6,2) R> A [,1] [,2] [,3] [1,] 1 3 5 [2,] 2 4 6
Example:
R> XY <- cbind(1:10,11:20) R> dim(XY) [1] 10 2
Accessing the values of an object
Example: display the third element of X and Y:
Example:
Example:
R> head(XY) [,1] [,2] [1,] 1 11 [2,] 2 12 [3,] 3 13 [4,] 4 14 [5,] 5 15 [6,] 6 16
Example:
R> colnames(A) <- c("a", "b", "c") R> A a b c [1,] 1 3 5 [2,] 2 4 6 R> A<-as.data.frame(A) R> A$a [1] 1 2
Operators
R> 2^2 [1] 4
Example
R> x <- 0.5 R> (0 < x) [1] TRUE R> x <- 1:3 R> y <- 1:3 R> (x == y) [1] TRUE TRUE TRUE
Example:
R> x<-1:6 R> y<-4:9 R> (x>5) [1] FALSE FALSE FALSE FALSE FALSE TRUE R> ! (x>5) [1] TRUE TRUE TRUE TRUE TRUE FALSE R> (x<3)&(y>4) [1] FALSE TRUE FALSE FALSE FALSE FALSE R> (x<5)&(y>4) [1] FALSE TRUE TRUE TRUE FALSE FALSE
A few functions
Example:
R> sum(X) [1] 15
Example:
R> cumsum(X) [1] 1 3 6 10 15
Example:
R> rowSums(XY) [1] 12 14 16 18 20 22 24 26 28 30 R> colSums(XY) [1] 55 155